BlueGenes development is at the point where we need to store BlueGenes specific data to a database. This is an important step because it paves the way for customisation, branding, and tool configuration, and an enhanced My Data section to let users manage all of their InterMine assets.
There are a few architecture and design decisions that need to be made now, and be made correctly. In particular: OAuth2 Authentication. If you’re up to speed on how InterMine and BlueGenes authenticate then feel free to skip to the bottom.
Background
The current InterMine web application is a monolith. Users login to the UI with a username and password and their identity gets stored in memory on the server (called the “session”). When they perform a query or upgrade a list the JSP code sends messages to the Java layer along with the user’s identity which is used to retrieve data from the object store and user profile.
For example, when Sally views her list page today, the workflow looks something like:
Figure 1

Everything you see in InterMine today lives somewhere layered between the JSP Web App and the Object Store.
BlueGenes works differently. It communicates with the Java layer, object store, and user profile entirely through web services known as the InterMine API. No exceptions. This cleaves the dependency between the visual tools that we develop and the lower level operations of InterMine such as handling queries.
When Sally views her list page in BlueGenes, the workflow looks more like this:
Figure 2

BlueGenes lives in the browser, not on the server. InterMine’s web services respond with raw data about her lists in JSON format and BlueGenes renders the page in the browser. This is equivalent to running Python scripts in your console to fetch your lists, resolve IDs, perform a search, etc.
Web services (InterMine or otherwise) are stateless by design. They can’t tell if requests are made by a new user or a revisiting one. In order for a web service to authorise a user the request must contain some sort of secret token as seen in Figure 2. Like any good web application, InterMine provides web services for authenticating a user and retrieving their identity token which can be used in future requests rather than a username and password.
BlueGenes Authentication
Now it gets a bit trickier. BlueGenes has its own small web server to provide the actual javascript application, and it requires database access to store BlueGenes specific information such as additional MyMine data, tool config, etc. It really looks more like this:
Figure 3
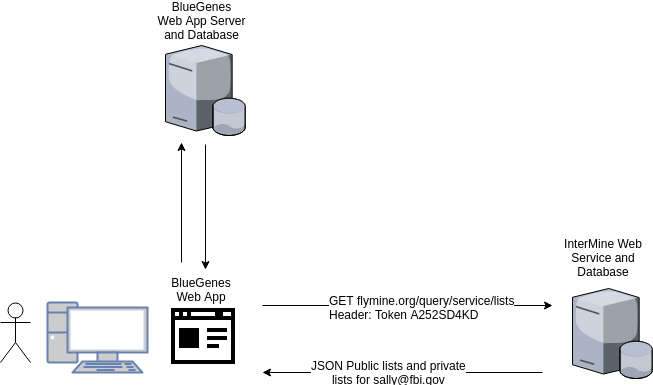
A user can authenticate using InterMine’s web services via the browser, but if they want to save user specific data to BlueGenes’s database using BlueGene’s web services then they need to provide an identity. BlueGenes does not have access to the user profile directly, so the authentication request needs to be piped through the BlueGenes server.
Figure 4
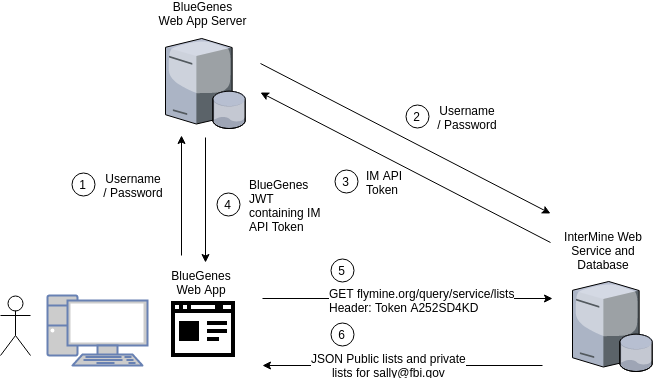
When Sally logs into BlueGenes she provides her username and password which is sent to the BlueGenes server rather than the InterMine server. If BlueGenes successfully authenticates as Sally then it sends her back her InterMine API token embedded in a signed JSON Web Token (JWT). All future requests between BlueGenes and InterMine will contain her API token, and all requests to the BlueGenes server will contain the signed JWT.
It sounds a bit complicated, but this only happens when logging in and remains hidden from the user. This configuration protects BlueGenes from storing passwords and doesn’t require direct access to the user profile.
The problem: OAuth2 Authentication
Logging into InterMine using your Google account uses the OAuth2 framework. For it to work you must configure Google’s developer console with a hardcoded URL that redirects users back to the application after they’ve authenticated. This redirection page is given a token that is exchanged by the servers for the user’s Google identity (email address and Google ID). We can do the same in BlueGenes:
- We put a Google Signin button in BlueGenes.
- Sally clicks it and is redirected to Google.
- Upon authentication Sally is sent back to BlueGenes with an authentication token.
- BlueGenes server exchanges the token for Sally’s Google ID.
So far so good. She can update her tool configurations and tags which are stored in the BlueGenes database.
Now Sally wants to save a list which is an action performed in InterMine, not BlueGenes. This requires an API token which she doesn’t yet have.
- She can’t authenticate with InterMine using a username and password because she doesn’t have one (she’s a Google user).
- She has no way of exchanging her Google ID with InterMine’s web services for an API token because InterMine has no way of trusting who she is. Anyone could access the end point and get a user’s API token if they knew their Google ID.
- BlueGenes can’t fetch her API token from the user profile because it doesn’t have access (by design).
There are a few workaround solutions but they couple BlueGenes to a single InterMine instance with varying degrees.
Solution 1: JWTs and sharing secrets
InterMine server gets a new end point that accepts a user ID and a JSON Web Token. The user’s API token is returned only if the signature on the JWT is valid.
Pain point: Both BlueGenes server and InterMine server will need matching secret keys. A third party cannot host their own BlueGenes and point it at a remote mine while supporting OAuth2 without knowing that mine’s secret key (aka access to all accounts).
InterMine admins could potentially whitelist third party instances of BlueGenes by generating secret keys for them, but this would be an active process of curation and still give third parties full access to all Google accounts..
Solution 2: Shared database
BlueGenes accesses the user profile directly.
Pain point: This requires database access which entirely rules out remote instances of BlueGenes
Solution 3: Double Login
InterMine has a URL redirect for Google authentication. It accepts a URL of a BlueGenes instance and generates a link with an embedded API key.
- A user clicks Google Login on BlueGenes and is redirected to Google
- After authenticating the user is redirected back to the BlueGenes server.
- BlueGenes generates a JWT containing the user’s identity.
- A mandatory button is then shown to “Authorise My Account to use Remote Data Sources” (which means InterMine server).
- Clicking the button sends the user to a /service/google-auth end point on the remote mine with a return_to parameters containing the URL of BlueGenes.
- The return_to parameter is stored in the session and the user is sent back to Google Login where they authorise for the second time.
- After authenticating the user is redirected to an InterMine /service/google-auth-redirect end point.
- The /service/google-auth-redirect page automatically redirects the user back to the BlueGenes URL stored in the session with the API token as a parameter
A workflow would look something like this:

There are quite a few steps, but steps 5+ are automatic.
Pain point: Users will have to double authentication the first time they login to Bluegenes, but we can make this as painless as possible. Also, if an admin is running both InterMine server and BlueGenes server then they’ll need two OAuth2 projects in their Google developer console (also a one time activity).
Solution 4: Outsource
We use a third party single sign-on vendor such as https://auth0.com/
Pain point: We can’t guarantee that InterMine admins will remain within the Terms of Service for their free offering to open source projects. Otherwise it’s very expensive.
Solution 3 seems to be the most feasible and keeps InterMine and BlueGenes completely decoupled. (Thanks, Yo!)
Does anyone feel strongly about a particular solution, or have other advice for bridging the OAuth2 gap? Feel free to leave a comment or join in the discussion on our mailing list (mailing list subscription link is here: https://lists.intermine.org/mailman/listinfo/dev)









You must be logged in to post a comment.